3 Best Practices for Writing R Scripts
This chapter is all about R scripts: what they are, when to use them, and how to write them—with specific emphasis making sure your scripts are easy to read, extend, debug, and reuse. Topics covered along the way include package management, how to use iteration, how to print or log output, and how to read input from the command line or from configuration files.
Learning Objectives
After completing this session, learners should be able to:
- Describe the advantages and disadvantages of using scripts
- Develop modular scripts that serve as applications and libraries
- Inspect and manage R’s software (package) environment
- Describe and use R’s for, while, and repeat loops
- Identify the most appropriate iteration strategy for a given task
- Explain strategies to organize iterative code
- Identify and explain the difference between R’s various printing functions
- Read user input from the command line into R
- Read data from configuration files into R
3.1 Introduction
An R script is a text file that contains R code. R scripts usually have the
extension .R. You might already use R scripts in your workflow (for example,
if you learned R from DataLab’s R Basics Workshop Series).
Even if you do, this chapter covers many different practices you can adopt to
use scripts more effectively. Although the chapter is focused on R, many of the
practices also apply to scripting other languages.
Notebooks, which usually have the extension .Rmd, are the main alternative to
scripts. Scripts and notebooks are both great ways to store code, and for most
projects, DataLab staff use a mix of both. To decide which one to use, think
about why you’re writing code. Compared to notebooks, the specific advantages
of scripts are that they are:
Simple. Scripts are text files and usually only contain one programming language (in our case, R). Notebooks require familiarity with Markdown, and some also require special software.
Easy to reuse. In R, you can run all of the code in a script with the
sourcefunction. This makes scripts a great way to create a library of reusable functions. Writing reusable scripts is also the first step towards developing packages to share with a wider audience.Easy to run in a command-line interface. R provides a command-line program,
Rscript, for running R scripts. Although you probably use a graphical interface to run code on your computer, high-performance computing environments do not always have a graphical interface, and even when they do, the default is usually a command-line interface. Running notebooks at the command-line is more difficult.
Scripts also have one big disadvantage compared to notebooks: results must be saved and viewed after the script runs, which means you have to plan ahead and write some extra code. As a result, we recommend scripts for code you plan to reuse or run at the command-line (especially if it will take a long time to run), and recommend notebooks for exploratory work. You can read more about how to decide in DataLab’s Reproducibility Principles and Practices reader.
This chapter is organized around a single case study (Section 3.2) that benefits from many of the practices we recommend. At a few points, the narrative branches off from the case study to take a closer look at a particular practice or topic in more general terms.
3.2 Case Study: U.S. Fruit Prices
The U.S. Department of Agriculture (USDA) Economic Research Service (ERS) computed national average retail prices for various fruits and vegetables in 2013, 2016, and most recently, 2020. These data provide partial insight into how much Americans must spend to eat a healthy diet with a variety of fruits and vegetables. The USDA ERS warns against using the data to compare prices across years because of differences in how products were categorized, but it can still be used to examine consumer costs in a given year.
The 2020 data are provided as per-fruit Microsoft Excel (.xlsx) files or as
an all-fruits comma-separated value (CSV) file ready to use with languages such
as R. Unfortunately, the 2016 and 2013 data were only distributed as per-fruit
files, and the structure of these files makes extracting the data non-trivial.
The goal of this case study is to develop an R script that can read the 2016 fruit data into a single all-fruit data frame. We’ll write the script in a way that makes it easy to extend to the 2016 vegetable data or the 2013 data.
3.2.1 Setting Up the Project
There are a few things you should do every time you start working on a new
project. First, create a new directory to store files related to the project.
This is usually called a repository or project directory. Give the
repository a descriptive name such as usda_fruit_prices.
The purpose of the repository is to keep files related to the project together, so that they’re easy to find and share. Choosing a descriptive name for the repository, and for files in general, will make it easier for others and future you to understand your work. You can read more about how to organize projects and choose names in DataLab’s Reproducibility Principles and Practices reader.
Next, create a data/ subdirectory in the repository to store data.
Subdirectories are a good way to keep repositories organized, especially if you
follow a naming convention. For instance, DataLab projects almost always have a
data/ subdirectory to store data.
Download the zipped 2016 fruit data and save it in the data/
subdirectory. Unzip the file with a zip program or R’s own unzip function.
Although the 2016 data include CSV files, these have the same structural
problems as the Excel files, so we’ll focus on the Excel files.
Finally, create an R/ subdirectory in the repository to store the project’s R
scripts, and make a new R script in subdirectory called extract_prices.R. You
can create and open the script with RStudio or your favorite text editor.
3.2.2 Setting Up the Script
Start every script with a few comments about its purpose, usage, inputs, and outputs. These comments will serve as a blueprint for your code and as documentation. Write them before you write any code—this will help you make your ideas more concrete and keep you focused when you do start writing code.
For extract_prices.R, the goal is to extract price data from the Excel file
for each fruit (or vegetable). We can extract the data to a data frame, since
the data are already in a tabular format. We can have the script save the data
frame as an RDS file, R’s native data format. If we want to do analysis on the
data, we can use a separate script that reads the RDS file. This is a
separation of concerns which ensures we can focus on the task at
hand—data extraction. Summarize these ideas in comments at the beginning of
extract_prices.R, by adding something like this:
# Extract a data frame of prices from USDA ERS fruit (and vegetable) price data
# Excel files. Save the data frame as an RDS file.The key to making scripts reusable is to break computational tasks into small steps and write a short function for each step. When another script (or project) involves a similar step, you can reuse or repurpose some of the functions. This function-focused, modular development strategy also makes it easier to find and fix bugs, since you can test each function on a variety of inputs to make sure it works as intended. The strategy may also help you tackle large and complex tasks, since the component steps should be smaller and simpler, and solving one provides positive feedback. Knowing what it means for a step to be “small” takes experience, but it’s usually better to err on the side of too small rather than too large. As a rule of thumb, a function with more than one screen of code is probably trying to do too much.
Scripts you plan to run, like extract_prices.R, need some kind of starting
point. Since we’re taking a function-focused approach, the starting point will
be a function. There’s no standard or conventional name for a starting point
function in R. Many other languages use a function called main as the
starting point, so we’ll copy that convention. Add a function called
main to extract_prices.R:
Now let’s think about the steps we need to take to extract the price data. We want to extract the data from many different Excel files. If you haven’t already, open a few of the files up in a spreadsheet program to take a look at the data. The first sheet of each file contains a table with the name of the fruit and prices sorted by how the fruit was prepared. The number of rows differs between fruits, but the overall structure doesn’t seem to change, which suggests we can read and process each file in the same way. There are two things you should do when you encounter a repetitive task like this one:
Start by focusing on a single case. We’ll start by focusing exclusively on extracting the data for pears (
pears 2016.xlsx).Once you get the code working on a single case, test that the code is general enough to handle other cases, and then use iteration to run the code on all cases. You’ll learn more about iteration later in this case study and in Section 3.4.
3.2.3 Reading the Pear Data
The first step to extracting the pear data is reading the pears 2016.xlsx
Excel file into R. We’ll use the read_excel function from the readxl
package to read the files, since R does not provide a built-in function to read
Excel files. Use the R prompt to install the package:
Do not add this code to the extract_prices.R script. In most cases, you
should not include code to install packages in your scripts. Instead, put
library calls and comments near the beginning of the script, to specify which
packages are required, and leave it to the user to install them. This prevents
needless, time-consuming reinstallation of packages.
We’re going to use the readxl package, so add a library call for the package
near the beginning of extract_prices.R, just after the purpose comments and
just before the main function:
# Extract a data frame of prices from USDA ERS fruit (and vegetable) price data
# Excel files. Save the data frame as an RDS file.
library("readxl")
# ...We’ll add more calls to library here as we decide to use more packages. This
way it’s easy for someone to see which packages are required as soon as they
open the script. Putting the calls to library in alphabetical order by
package name is also a good idea. You can read more about how to manage the
packages a script or project requires in Section 3.3.
When developing a script, it’s often helpful to run the code, check what’s
working, and make changes, repeating this process until the script is finished.
Fortunately, R provides a convenient way to run an entire script: the source
function. The first argument is the path to the script you want to run. In your
R prompt, try sourcing extract_prices.R:
The path is relative to R’s working directory; see this
section of DataLab’s R Basics reader for details about
how to change the working directory. The code above assumes the repository is
the working directory. If the script was sourced correctly, R will load the
readxl package and define a function main, which currently does nothing. As
you add code to main and add other functions, you can source the script again
to update these functions in your R session. If you’re using R Studio, you can
also check the “source on save” box in the graphical user interface to make R
automatically call source every time you save the script. The source
function can save you time during development, but of course it’s also okay to
run code by typing or pasting it into the prompt.
Take a look at the help file for the read_excel function (run ?read_excel
in the R prompt). The first argument is the path to the Excel file to read.
Change the code in the main function to read the pear data:
Source the script and then call main in the R prompt:
## New names:
## • `` -> `...2`
## • `` -> `...3`
## • `` -> `...4`
## • `` -> `...5`
## • `` -> `...6`
## • `` -> `...7`## # A tibble: 13 × 7
## Pears—Average retail price per pound an…¹ ...2 ...3 ...4 ...5 ...6 ...7
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Form Aver… <NA> Prep… Size… <NA> Aver…
## 2 <NA> <NA> <NA> yiel… cup … <NA> per …
## 3 Fresh1 1.51… per … 0.9 0.36… poun… 0.61…
## 4 Canned <NA> <NA> <NA> <NA> <NA> <NA>
## 5 Packed in juice2 1.78… per … 1 0.54… poun… 0.96…
## 6 Packed in syrup or water3 1.57… per … 0.65 0.44… poun… 1.06…
## 7 1The USDA National Nutrient Database for… <NA> <NA> <NA> <NA> <NA> <NA>
## 8 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 9 2Consumers are assumed to eat the solid … <NA> <NA> <NA> <NA> <NA> <NA>
## 10 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 11 3The syrup (or water) is discarded prior… <NA> <NA> <NA> <NA> <NA> <NA>
## 12 <NA> <NA> <NA> <NA> <NA> <NA> <NA>
## 13 Source: Calculated by USDA, Economic Res… <NA> <NA> <NA> <NA> <NA> <NA>
## # ℹ abbreviated name:
## # ¹`Pears—Average retail price per pound and per cup equivalent, 2016`This returns the data in the Excel file, but there’s still a lot of cleanup to do. Since we’re eventually going to extract data about many different fruits, let’s start by getting the name of the fruit from the Excel file.
3.2.4 Getting the Fruit Name
Notice that the name of each fruit is in the top left cell (A1) of its file.
The read_excel function treats the first row of cells as headers, so in the
data frame, the name of the fruit is in the first column name. Create a
function called get_fruit_name to get this column name:
Also update the main function to save the data frame in a variable and call
get_fruit_name:
main = function() {
sheet = read_excel("data/fruit/pears 2016.xlsx")
get_fruit_name(sheet)
# More to come...
}Once again, source the script and call main. The return value will be a
string:
## [1] "Pears—Average retail price per pound and per cup equivalent, 2016"This has the name of the fruit, but also some extra text. To get just the name, we need to do some string processing. We’ll use the stringr package for string processing.
Install the package and add it to the library calls at the
beginning of extract_prices.R:
# Extract a data frame of prices from USDA ERS fruit (and vegetable) price data
# Excel files. Save the data frame as an RDS file.
library("readxl")
library("stringr")
# ...We’ll use several functions from stringr, but each will be explained in context. The package provides detailed documentation as well as a cheatsheet. If you want to learn more about string processing and stringr in general, see Chapter 1 of this reader.
The name of the fruit is separated from the extra text by an em dash (the —
character, which is longer than minus, the - character). We can use the
str_split_fixed function to split the text into two parts at the em dash. The
function’s first argument is the string to split, its second is the character
on which to split (copy and paste the em dash), and its third is the number of
parts. The function returns a matrix where each column corresponds to one of
the split parts. In extract_prices.R, update get_fruit_name to split the
text and get the first element, the fruit name:
As usual, source the script and call main. The result is just the fruit
name:
## [1] "Pears"We’ll use the fruit name later to label the data from each file.
3.2.5 Getting the Column Names
The column names on the sheet data frame come from the first row of the file,
but the actual headers of the table are in the second and third rows of the
file (rows 1 and 2 of the data frame):
## # A tibble: 6 × 7
## Pears—Average retail price per pound and…¹ ...2 ...3 ...4 ...5 ...6 ...7
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Form Aver… <NA> Prep… Size… <NA> Aver…
## 2 <NA> <NA> <NA> yiel… cup … <NA> per …
## 3 Fresh1 1.51… per … 0.9 0.36… poun… 0.61…
## 4 Canned <NA> <NA> <NA> <NA> <NA> <NA>
## 5 Packed in juice2 1.78… per … 1 0.54… poun… 0.96…
## 6 Packed in syrup or water3 1.57… per … 0.65 0.44… poun… 1.06…
## # ℹ abbreviated name:
## # ¹`Pears—Average retail price per pound and per cup equivalent, 2016`Let’s create another function, get_column_names, to convert these rows into a
vector we can use as column names. Add the function to extract_prices.R, with
a code to get rows 1 and 2 of the data frame:
As usual, update main to call the new function:
main = function() {
sheet = read_excel("data/fruit/pears 2016.xlsx")
fruit = get_fruit_name(sheet)
get_column_names(sheet)
# More to come...
}Then source extract_prices.R and call main:
## # A tibble: 2 × 7
## Pears—Average retail price per pound and…¹ ...2 ...3 ...4 ...5 ...6 ...7
## <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Form Aver… <NA> Prep… Size… <NA> Aver…
## 2 <NA> <NA> <NA> yiel… cup … <NA> per …
## # ℹ abbreviated name:
## # ¹`Pears—Average retail price per pound and per cup equivalent, 2016`For each column, we need to paste the rows together. This “for each” is a hint
that the task that can be solved with iteration. Before proceeding, take a
moment to learn more about iteration by reading through Section
3.4. For this task, the result should be one string per
column, so you can use sapply to iterate over the columns.
By setting its collapse argument, you can use R’s built-in paste function
to paste together the elements of a vector together. The value of collapse is
inserted between the elements. For the fruit price headers, there should be a
space between the row text, so we’ll set collapse = " ". In
extract_prices.R, update get_column_names, source the file, and call
main:
get_column_names = function(sheet) {
# Get rows 1 and 2, all columns.
names = sheet[1:2, ]
sapply(names, paste, collapse = " ")
}## Pears—Average retail price per pound and per cup equivalent, 2016
## "Form NA"
## ...2
## "Average retail price NA"
## ...3
## "NA NA"
## ...4
## "Preparation yield factor"
## ...5
## "Size of a cup equivalent"
## ...6
## "NA NA"
## ...7
## "Average price per cup equivalent"The result is almost right, but the paste function converts all of the
missing values NA to literal text "NA". To fix this, replace all of the
missing values with the empty string "" before pasting the rows together.
Let’s add a call to str_trim at the end of get_column_names to trim off any
excess spaces as well. Once again, update the function in extract_prices.R,
source the file, and call main:
get_column_names = function(sheet) {
# Get rows 1 and 2, all columns.
names = sheet[1:2, ]
# Paste together, treating NAs as empty string.
names[is.na(names)] = ""
names = sapply(names, paste, collapse = " ")
# Trim off extra whitespace.
str_trim(names)
}## [1] "Form" "Average retail price"
## [3] "" "Preparation yield factor"
## [5] "Size of a cup equivalent" ""
## [7] "Average price per cup equivalent"Notice that now two of the elements are just the empty string "". Take a look
at the Excel file. Can you see why this happens? These are the two columns with
units, so let’s call them units1 and units2. The numbers are to make the
column names are unique. Let’s also make the other column names easier to use
by converting them to lowercase with str_to_lower and replacing the spaces
with underscores _ with str_replace_all. In extract_prices.R, update
get_column_names, source the file, and call main:
get_column_names = function(sheet) {
# Get rows 1 and 2, all columns.
names = sheet[1:2, ]
# Paste together, treating NAs as empty strings.
names[is.na(names)] = ""
names = sapply(names, paste, collapse = " ")
# Trim off extra whitespace.
names = str_trim(names)
# Replace empty names with "unitsN"
is_empty = names == ""
n_empty = sum(is_empty)
names[is_empty] = paste("units", seq_len(n_empty), sep = "")
# Convert to lowercase and replace spaces with underscores.
names = str_to_lower(names)
str_replace_all(names, " +", "_")
}## [1] "form" "average_retail_price"
## [3] "units1" "preparation_yield_factor"
## [5] "size_of_a_cup_equivalent" "units2"
## [7] "average_price_per_cup_equivalent"These names look good enough to use.
3.2.6 Getting the Price Data
With the fruit name and column names taken care of, now it’s time to get the
actual price data. There are a few different ways to identify the rows with
price data. One way is to look for entries in the second column. Let’s set up a
new function, get_fruit_prices. Eventually we’ll make this function return
the price data, but to get started, let’s make it return the indexes of the
first and last row with entries in the second column. Add this code to
extract_prices.R:
Then update main, source the file, and call main:
main = function() {
sheet = read_excel("data/fruit/pears 2016.xlsx")
fruit = get_fruit_name(sheet)
col_names = get_column_names(sheet)
get_fruit_prices(sheet)
}## [1] 1 6The first index corresponds to the header row, but we already dealt with the header in Section 3.2.5. Since the header spans two rows, we can skip it by adding 2 to the first index. The second index corresponds to the last row of prices, so it seems like this is an effective strategy for getting the price data (we can’t be completely sure until we test the strategy on other fruits).
Update get_fruit_prices and main in extract_prices.R to get the subset of
rows with price data and to change the column names to the ones we computed
earlier:
get_fruit_prices = function(sheet, col_names) {
# Get all rows from first entry to last entry in column 2.
is_filled = !is.na(sheet[, 2])
idx = range(which(is_filled))
prices = sheet[seq(idx[1] + 2, idx[2]), ]
# Set column names.
names(prices) = col_names
prices
}
main = function() {
sheet = read_excel("data/fruit/pears 2016.xlsx")
fruit = get_fruit_name(sheet)
col_names = get_column_names(sheet)
get_fruit_prices(sheet, col_names)
}## # A tibble: 4 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## <chr> <chr> <chr> <chr>
## 1 Fresh1 1.5175920102 per pou… 0.9
## 2 Canned <NA> <NA> <NA>
## 3 Packed in juice2 1.7876110202 per pou… 1
## 4 Packed in syrup or water3 1.5716448654999999 per pou… 0.65
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>The result looks good, but notice that the "Canned" row has a several missing
values. Take a look at the original Excel file to figure out why this happens.
The "Canned" row is the beginning of a subgroup of prices for canned fruit.
The data frame returned by get_fruit_prices will be more convenient for
analysis if we exclude the "Canned" row and instead mark the following two
rows as canned. For instance, we might label their form as "Canned, packed in juice" and "Canned, packed in syrup or water".
One way to approach the subgroup labeling task is to compute the subgroup for
each row, then use the split-apply pattern to process each
subgroup. The "Canned" row doesn’t have an entry in the second column, so
maybe we can use that to detect where each subgroup begins. You can use the
is.na to find those rows, and the cumulative sum function cumsum to make
the subgroup labels (since FALSE counts as 0 and TRUE counts as 1).
Update get_fruit_prices in extract_prices.R to split the price data by
subgroup labels, source the file, and call main:
get_fruit_prices = function(sheet, col_names) {
# Get all rows from first entry to last entry in column 2.
is_filled = !is.na(sheet[, 2])
idx = range(which(is_filled))
prices = sheet[seq(idx[1] + 2, idx[2]), ]
# Set column names.
names(prices) = col_names
# Compute form subgroups.
subgroups = cumsum(is.na(prices[, 2]))
split(prices, subgroups)
}## $`0`
## # A tibble: 1 × 7
## form average_retail_price units1 preparation_yield_factor
## <chr> <chr> <chr> <chr>
## 1 Fresh1 1.5175920102 per pound 0.9
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>
##
## $`1`
## # A tibble: 3 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## <chr> <chr> <chr> <chr>
## 1 Canned <NA> <NA> <NA>
## 2 Packed in juice2 1.7876110202 per pou… 1
## 3 Packed in syrup or water3 1.5716448654999999 per pou… 0.65
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>The result is a separate data frame for each subgroup. For each subgroup, we
need to check whether the first row has an entry in the second column. If it
doesn’t, then the first row is a group name (like "Canned") that should be
pasted onto the other rows in the group. Add a function
process_price_subgroup to extract_prices.R to do this for a single group:
process_price_subgroup = function(subgroup) {
if (is.na(subgroup[1, 2])) {
# First row is group name, so remove the row and paste the group name onto
# the other rows.
name = subgroup[[1, 1]]
subgroup = subgroup[-1, ]
form = str_to_lower(str_trim(subgroup$form))
subgroup$form = paste(name, form, sep = ", ")
}
subgroup
}You can use lapply to apply this function to each subgroup, and do.call
with rbind to recombine the data frames. Update get_fruit_prices in
extract_prices.R, source the file, and call main:
get_fruit_prices = function(sheet, col_names) {
# Get all rows from first entry to last entry in column 2.
is_filled = !is.na(sheet[, 2])
idx = range(which(is_filled))
prices = sheet[seq(idx[1] + 2, idx[2]), ]
# Set column names.
names(prices) = col_names
# Compute form subgroups.
subgroups = cumsum(is.na(prices[, 2]))
by_subgroup = split(prices, subgroups)
# Process each subgroup, then combine them into a single data frame.
result = lapply(by_subgroup, process_price_subgroup)
do.call(rbind, result)
}## # A tibble: 3 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## * <chr> <chr> <chr> <chr>
## 1 Fresh1 1.5175920102 per p… 0.9
## 2 Canned, packed in juice2 1.7876110202 per p… 1
## 3 Canned, packed in syrup or… 1.5716448654999999 per p… 0.65
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>It seems like the script now works for single fruits!
3.2.7 Testing the Script
Let’s try a different fruit, peaches, to make
sure the script is sufficiently general. In extract_prices.R, change the file
loaded in main, source the script, and call main:
main = function() {
sheet = read_excel("data/fruit/peaches 2016.xlsx")
fruit = get_fruit_name(sheet)
col_names = get_column_names(sheet)
get_fruit_prices(sheet, col_names)
}## # A tibble: 4 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## * <chr> <chr> <chr> <chr>
## 1 Fresh1 1.6779315817 per p… 0.96
## 2 Canned, packed in juice2 1.8800453697999999 per p… 1
## 3 Canned, packed in syrup or… 1.5862877904999999 per p… 0.65
## 4 Canned, frozen 3.1870806846000002 per p… 1
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>Uh-oh, the last row is "Canned, frozen", which doesn’t seem correct. Take a
look at the Excel file for peaches to see what went wrong. The bug is that we
assumed subgroups are always separated by header rows where there’s no price
(like the "Canned" row). In the peaches file, the row with prices for frozen
peaches is not separated this way. Looking at a few more of the files, it seems
like there are only a few subgroups: Canned, Dried, Fresh, Frozen, and
Juice. So let’s change the script to check specifically for these.
You can use stringr’s str_starts function to detect entries in the form
column which start with one of the subgroup names. The second argument to the
function is a regular expression (see Section 1.4.2), a
string which describes a pattern. You can use the pipe character | in a
regular expression to separate several different options. Update the code for
get_fruit_prices in extract_prices.R, source the file, and call main:
get_fruit_prices = function(sheet, col_names) {
# Get all rows from first entry to last entry in column 2.
is_filled = !is.na(sheet[, 2])
idx = range(which(is_filled))
prices = sheet[seq(idx[1] + 2, idx[2]), ]
# Set column names.
names(prices) = col_names
# Compute form subgroups.
subgroup_names = "Canned|Dried|Fresh|Frozen|Juice"
subgroups = cumsum(str_starts(prices$form, subgroup_names))
by_subgroup = split(prices, subgroups)
# Process each subgroup, then combine them into a single data frame.
result = lapply(by_subgroup, process_price_subgroup)
do.call(rbind, result)
}## # A tibble: 4 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## * <chr> <chr> <chr> <chr>
## 1 Fresh1 1.6779315817 per p… 0.96
## 2 Canned, packed in juice2 1.8800453697999999 per p… 1
## 3 Canned, packed in syrup or… 1.5862877904999999 per p… 0.65
## 4 Frozen 3.1870806846000002 per p… 1
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>This appears to be correct. Test the script on the pears data again as well, to make sure the changes didn’t introduce other bugs:
## # A tibble: 3 × 7
## form average_retail_price units1 preparation_yield_fa…¹
## * <chr> <chr> <chr> <chr>
## 1 Fresh1 1.5175920102 per p… 0.9
## 2 Canned, packed in juice2 1.7876110202 per p… 1
## 3 Canned, packed in syrup or… 1.5716448654999999 per p… 0.65
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 3 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>Some of the columns in the data frame have inappropriate data types. This can be fixed with just a few lines of code, but since it doesn’t provide any additional insight into how to write scripts, we won’t do so here. If you want to know more about how to fix “messy data” problems, read Chapter 1.
3.2.8 Iterating Over All Fruit
The final step in extracting the fruit price data is to modify
extract_prices.R to extract the prices of all fruits. This is also a good
time to make several small changes to the script that will make it more
convenient to use.
First, change the name of main to read_price_file, make the path to the
Excel file a parameter called path, and limit the Excel sheet to seven
columns:
read_price_file = function(path) {
sheet = read_excel(path)
sheet = sheet[, 1:7]
fruit = get_fruit_name(sheet)
col_names = get_column_names(sheet)
prices = get_fruit_prices(sheet, col_names)
prices$fruit = fruit
prices
}This change makes it possible to reuse the function for many different fruit (and vegetable) Excel files.
Next, let’s create a new main function, which will call read_price_file on
all of the Excel files in a user-specified directory. The function should have
two parameters: input_dir, for the path to the directory that contains the
Excel files, and output_path, for a path to an RDS file where the resulting
data frame should be saved. You can use list.files to get a vector of paths
to all files in the input directory, lapply to iterate over the files, and
saveRDS to save the result. Here’s the code to add to extract_prices.R:
main = function(
input_dir = "data/fruit/",
output_path = "prices.rds"
) {
# Get vector of all files in directory.
message("Input directory: ", input_dir)
paths = list.files(input_dir, pattern = "*.xlsx", full.names = TRUE)
# Read each file and combine the resulting data frames.
result = lapply(paths, function(path) {
message("Reading: ", path)
read_price_file(path)
})
result = do.call(rbind, result)
# Save the result to an RDS file.
saveRDS(result, output_path)
message("Wrote:", output_path)
result
}The code for the function includes several calls to message to print
informational messages. It’s a good idea to include these so that you (or other
users) can verify that the paths to files are correct and can quickly diagnose
any bugs you encounter. You can read more about various ways to print output in
R in Section 3.5.
Once again, test that the code works by sourcing the script and calling main:
## # A tibble: 62 × 8
## form average_retail_price units1 preparation_yield_fa…¹
## <chr> <chr> <chr> <chr>
## 1 Fresh1 1.6155336441000001 per p… 0.9
## 2 Applesauce2 1.0491006433000001 per p… 1
## 3 Juice, ready to drink3 0.63113252779999995 per p… 1
## 4 Frozen4 0.51046574550000001 per p… 1
## 5 Fresh1 3.0871378169999999 per p… 0.93
## 6 Canned, packed in juice2 1.4742760156000001 per p… 1
## 7 Canned, packed in syrup, … 1.8552908088 per p… 0.65
## 8 Dried4 7.3309645323000003 per p… 1
## 9 Fresh1 0.54941729279999996 per p… 0.64
## 10 Frozen1 3.63694066 per p… 1
## # ℹ 52 more rows
## # ℹ abbreviated name: ¹preparation_yield_factor
## # ℹ 4 more variables: size_of_a_cup_equivalent <chr>, units2 <chr>,
## # average_price_per_cup_equivalent <chr>, fruit <chr>This looks almost perfect. There are still a few minor issues with data types, and also a small bug (related to frozen juice), but all of these can be fixed with small changes to the script.
3.3 Managing Environments
Your computing environment is the collection of hardware and software you use to run code. Code developed for one computing environment will not necessarily run correctly in another. Scripting languages like R and Python are designed to work with a wide variety of hardware, so in the context of these languages, software, especially package versions, are the primary cause of incompatibility.
Documenting the computing environment your code was designed for is an important part of making your research (or other work) reproducible. One way to do this is to maintain documentation that lists the specific packages your code depends on, with complete version information.
You can get version information about your R environment with the sessionInfo
function:
## R version 4.3.2 (2023-10-31)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Arch Linux
##
## Matrix products: default
## BLAS: /usr/lib/libblas.so.3.12.0
## LAPACK: /usr/lib/liblapack.so.3.12.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: US/Pacific
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] stringr_1.5.1 readxl_1.4.3
##
## loaded via a namespace (and not attached):
## [1] vctrs_0.6.5 cli_3.6.2 knitr_1.45 rlang_1.1.2
## [5] xfun_0.41 stringi_1.8.3 jsonlite_1.8.8 glue_1.6.2
## [9] htmltools_0.5.7 sass_0.4.8 fansi_1.0.6 rmarkdown_2.25
## [13] cellranger_1.1.0 evaluate_0.23 jquerylib_0.1.4 tibble_3.2.1
## [17] fastmap_1.1.1 yaml_2.3.8 lifecycle_1.0.4 bookdown_0.37
## [21] compiler_4.3.2 pkgconfig_2.0.3 digest_0.6.34 R6_2.5.1
## [25] utf8_1.2.4 pillar_1.9.0 magrittr_2.0.3 bslib_0.6.1
## [29] tools_4.3.2 cachem_1.0.8Documentation is often neglected, and it’s easy to forget that you updated a package, so you might prefer a more automated approach to recording your R environment. The renv package provides functions to record and recreate environments. See the package documentation for details about how to use the package.
3.4 Iteration Strategies
R is powerful tool for automating tasks that have repetitive steps. For example, you can:
- Apply a transformation to an entire column of data.
- Compute distances between all pairs from a set of points.
- Read a large collection of files from disk in order to combine and analyze the data they contain.
- Simulate how a system evolves over time from a specific set of starting parameters.
- Scrape data from many pages of a website.
You can implement concise, efficient solutions for these kinds of tasks in R by using iteration, which means repeating a computation many times. R provides four different strategies for writing iterative code:
- Vectorization, where a function is implicitly called on each element of a vector. See this section of DataLab’s R Basics reader for more details.
- Apply functions, where a function is explicitly called on each element of a vector or array. See this section of DataLab’s R Basics reader for more details.
- Loops, where an expression is evaluated repeatedly until some condition is met.
- Recursion, where a function calls itself.
Vectorization is the most efficient and most concise iteration strategy, but also the least flexible, because it only works with vectorized functions and vectors. Apply functions are more flexible—they work with any function and any data structure with elements—but less efficient and less concise. Loops and recursion provide the most flexibility but are the least concise. In recent versions of R, apply functions and loops are similar in terms of efficiency. Recursion tends to be the least efficient iteration strategy in R.
The rest of this section explains how to write loops and how to choose which iteration strategy to use. We assume you’re already comfortable with vectorization and have at least some familiarity with apply functions.
3.4.1 For-loops
A for-loop evaluates an expression once for each element of a vector or
list. The for keyword creates a for-loop. The syntax is:
The variable I is called an induction variable. At the beginning of each
iteration, I is assigned the next element of DATA. The loop iterates once
for each element, unless a keyword instructs R to exit the loop early (more
about this in Section 3.4.4). As with if-statements and functions,
the curly braces { } are only required if the body contains multiple lines of
code. Here’s a simple for-loop:
## Hi from iteration 1## Hi from iteration 2## Hi from iteration 3## Hi from iteration 4## Hi from iteration 5## Hi from iteration 6## Hi from iteration 7## Hi from iteration 8## Hi from iteration 9## Hi from iteration 10When some or all of the iterations in a task depend on results from prior iterations, loops tend to be the most appropriate iteration strategy. For instance, loops are a good way to implement time-based simulations or compute values in recursively defined sequences.
As a concrete example, suppose you want to compute the result of starting from the value 1 and composing the sine function 100 times:
## [1] 0.1688525Unlike other iteration strategies, loops don’t return a result automatically. It’s up to you to use variables to store any results you want to use later. If you want to save a result from every iteration, you can use a vector or a list indexed on the iteration number:
n = 1 + 100
result = numeric(n)
result[1] = 1
for (i in 2:n) {
result[i] = sin(result[i - 1])
}
plot(result)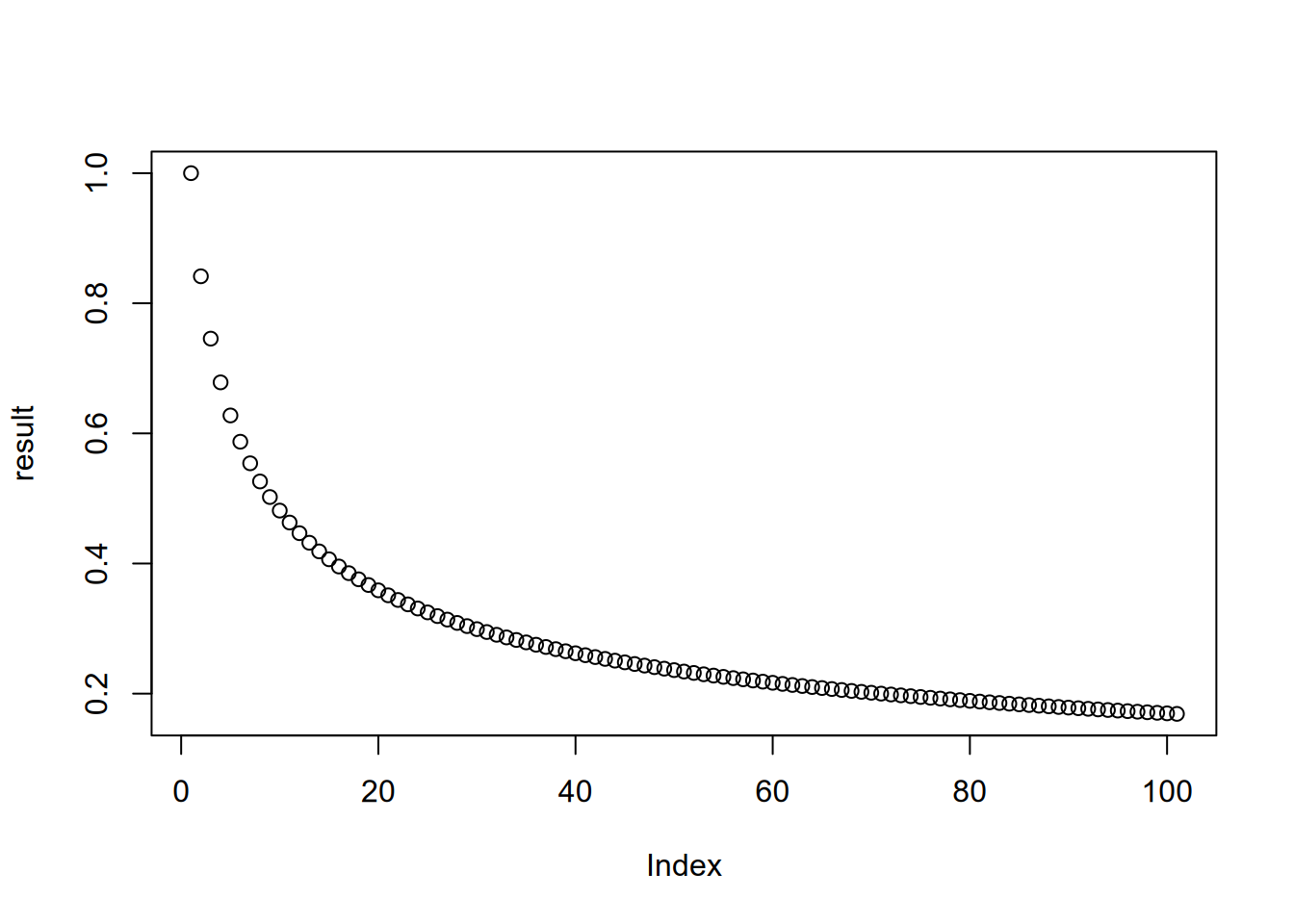
Section 3.4.3 explains this in more detail.
If the iterations in a task are not dependent, it’s preferable to use vectorization or apply functions instead of a loop. Vectorization is more efficient, and apply functions are usually more concise.
In some cases, you can use vectorization to handle a task even if the
iterations are dependent. For example, you can use vectorized exponentiation
and the sum function to compute the sum of the cubes of many numbers:
## [1] 10019103.4.2 While-loops
A while-loop runs a block of code repeatedly as long as some condition is
TRUE. The while keyword creates a while-loop. The syntax is:
The CONDITION should be a scalar logical value or an expression that returns
one. At the beginning of each iteration, R checks the CONDITION and exits the
loop if it’s FALSE. As always, the curly braces { } are only required if
the body contains multiple lines of code. Here’s a simple while-loop:
## Hello from iteration 1## Hello from iteration 2## Hello from iteration 3## Hello from iteration 4## Hello from iteration 5## Hello from iteration 6## Hello from iteration 7## Hello from iteration 8## Hello from iteration 9## Hello from iteration 10Notice that this example does the same thing as the simple for-loop in Section 3.4.1, but requires 5 lines of code instead of 2. While-loops are a generalization of for-loops, and only do the bare minimum necessary to iterate. They tend to be most useful when you don’t know how many iterations will be necessary to complete a task.
As an example, suppose you want to add up the integers in order until the total is greater than 50:
total = 0
i = 1
while (total < 50) {
total = total + i
message("i is ", i, " total is ", total)
i = i + 1
}## i is 1 total is 1## i is 2 total is 3## i is 3 total is 6## i is 4 total is 10## i is 5 total is 15## i is 6 total is 21## i is 7 total is 28## i is 8 total is 36## i is 9 total is 45## i is 10 total is 55## [1] 55## [1] 113.4.3 Saving Multiple Results
Loops often produce a different result for each iteration. If you want to save more than one result, there are a few things you must do.
First, set up an index vector. The index vector should usually correspond to
the positions of the elements in the data you want to process. The seq_along
function returns an index vector when passed a vector or list. For instance:
The loop will iterate over the index rather than the input, so the induction variable will track the current iteration number. On the first iteration, the induction variable will be 1, on the second it will be 2, and so on. Then you can use the induction variable and indexing to get the input for each iteration.
Second, set up an empty output vector or list. This should usually also correspond to the input, or one element longer (the extra element comes from the initial value). R has several functions for creating vectors:
logical,integer,numeric,complex, andcharactercreate an empty vector with a specific type and lengthvectorcreates an empty vector with a specific type and lengthrepcreates a vector by repeating elements of some other vector
Empty vectors are filled with FALSE, 0, or "", depending on the type of
the vector. Here are some examples:
## [1] FALSE FALSE FALSE## [1] 0 0 0 0## [1] 1 2 1 2Let’s create an empty numeric vector congruent to the numbers vector:
As with the input, you can use the induction variable and indexing to set the output for each iteration.
Creating a vector or list in advance to store something, as we’ve just done, is
called preallocation. Preallocation is extremely important for efficiency
in loops. Avoid the temptation to use c or append to build up the output
bit by bit in each iteration.
Finally, write the loop, making sure to get the input and set the output. As an
example, this loop adds each element of numbers to a running total and
squares the new running total:
for (i in index) {
prev = if (i > 1) result[i - 1] else 0
result[i] = (numbers[i] + prev)^2
}
result## [1] 1.000000e+00 4.840000e+02 2.371690e+05 5.624534e+10 3.163538e+213.4.4 Break & Next
The break keyword causes a loop to immediately exit. It only makes sense to
use break inside of an if-statement.
For example, suppose you want to print each string in a vector, but stop at the
first missing value. You can do this with a for-loop and the break keyword:
my_messages = c("Hi", "Hello", NA, "Goodbye")
for (msg in my_messages) {
if (is.na(msg))
break
message(msg)
}## Hi## HelloThe next keyword causes a loop to immediately go to the next iteration. As
with break, it only makes sense to use next inside of an if-statement.
Let’s modify the previous example so that missing values are skipped, but don’t cause printing to stop. Here’s the code:
## Hi## Hello## GoodbyeThese keywords work with both for-loops and while-loops.
3.4.5 Planning for Iteration
At first it may seem difficult to decide if and what kind of iteration to use. Start by thinking about whether you need to do something over and over. If you don’t, then you probably don’t need to use iteration. If you do, then try iteration strategies in this order:
- Vectorization
- Apply functions
- Try an apply function if iterations are independent.
- Loops
- Try a for-loop if some iterations depend on others.
- Try a while-loop if the number of iterations is unknown.
- Recursion (which isn’t covered here)
- Convenient for naturally recursive tasks (like Fibonacci), but often there are faster solutions.
Start by writing the code for just one iteration. Make sure that code works; it’s easy to test code for one iteration.
When you have one iteration working, then try using the code with an iteration
strategy (you will have to make some small changes). If it doesn’t work, try to
figure out which iteration is causing the problem. One way to do this is to use
message to print out information. Then try to write the code for the broken
iteration, get that iteration working, and repeat this whole process.
3.4.6 Case Study: The Collatz Conjecture
The Collatz Conjecture is a conjecture in math that was introduced in 1937 by Lothar Collatz and remains unproven today, despite being relatively easy to explain. Here’s a statement of the conjecture:
Start from any positive integer. If the integer is even, divide by 2. If the integer is odd, multiply by 3 and add 1.
If the result is not 1, repeat using the result as the new starting value.
The result will always reach 1 eventually, regardless of the starting value.
The sequences of numbers this process generates are called Collatz
sequences. For instance, the Collatz sequence starting from 2 is 2, 1. The
Collatz sequence starting from 12 is 12, 6, 3, 10, 5, 16, 8, 4, 2, 1.
You can use iteration to compute the Collatz sequence for a given starting value. Since each number in the sequence depends on the previous one, and since the length of the sequence varies, a while-loop is the most appropriate iteration strategy:
n = 5
i = 0
while (n != 1) {
i = i + 1
if (n %% 2 == 0) {
n = n / 2
} else {
n = 3 * n + 1
}
message(n, " ", appendLF = FALSE)
}## 16 8 4 2 1As of 2020, scientists have used computers to check the Collatz sequences for every number up to approximately \(2^{64}\). For more details about the Collatz Conjecture, check out this video.
For another example, see Liza Wood’s Real-world Function Writing Mini-reader.
3.5 Printing Output
This section introduces several different functions for printing output and making that output easier to read.
3.5.1 The print Function
The print function prints a string representation of an object to the
console. The string representation is usually formatted in a way that exposes
details important to programmers rather than users.
For example, when printing a vector, the function prints the position of the
first element on each line in square brackets [ ]:
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100The print function also prints quotes around strings:
## [1] "Hi"These features make the print function ideal for printing information when
you’re trying to understand some code or diagnose a bug. On the other hand,
these features also make print a bad choice for printing output or status
messages for users (including you).
R calls the print function automatically anytime a result is returned at the
prompt. Thus it’s not necessary to call print to print something when you’re
working directly in the console—only from within loops, functions, scripts,
and other code that runs non-interactively.
The print function is an S3 generic (see Section 6.4), so you if you
create an S3 class, you can define a custom print method for it. For S4
objects, R uses the S4 generic show instead of print.
3.5.2 The message Function
To print output for users, the message function is the one you should use.
The main reason for this is that the message function is part of R’s
conditions system for reporting status information as code runs. This makes
it easier for other code to detect, record, respond to, or suppress the output
(see Section 4.2 to learn more about R’s conditions
system).
The message function prints its argument(s) and a newline to the console:
## Hello world!If an argument isn’t a string, the function automatically and silently attempts to coerce it to one:
## 4Some types of objects can’t be coerced to a string:
## Error in FUN(X[[i]], ...): cannot coerce type 'builtin' to vector of type 'character'For objects with multiple elements, the function pastes together the string representations of the elements with no separators in between:
## 123Similarly, if you pass the message function multiple arguments, it pastes
them together with no separators:
## Hi, my name is R and x is 123This is a convenient way to print names or descriptions alongside values from
your code without having to call a formatting function like paste.
You can make the message function print something without adding a newline at
the end by setting the argument appendLF = FALSE. The difference can be easy
to miss unless you make several calls to message, so the say_hello function
in this example calls message twice:
say_hello = function(appendLF) {
message("Hello", appendLF = appendLF)
message(" world!")
}
say_hello(appendLF = TRUE)## Hello## world!## Hello world!Note that RStudio always adds a newline in front of the prompt, so making an
isolated call to message with appendLF = FALSE appears to produce the same
output as with appendLF = TRUE. This is an example of a situation where
RStudio leads you astray: in an ordinary R console, the two are clearly
different.
3.5.3 The cat Function
The cat function, whose name stands for “concatenate and print,” is a
low-level way to print output to the console or a file. The message function
prints output by calling cat, but cat is not part of R’s conditions system.
The cat function prints its argument(s) to the console. It does not add a
newline at the end:
## HelloAs with message, RStudio hides the fact that there’s no newline if you make
an isolated call to cat.
The cat function coerces its arguments to strings and concatenates them. By
default, a space is inserted between arguments and their elements:
## 4## 1 2 3## Hello NickYou can set the sep parameter to control the separator cat inserts:
## Hello|world|1|2|3If you want to write output to a file rather than to the console, you can call
cat with the file parameter set. However, it’s preferable to use functions
tailored to writing specific kinds of data, such as writeLines (for text) or
write.table (for tabular data), since they provide additional options to
control the output.
Many scripts and packages still use cat to print output, but the message
function provides more flexibility and control to people running the code. Thus
it’s generally preferable to use message in new code. Nevertheless, there are
a few specific cases where cat is useful—for example, if you want to pipe
data to a UNIX shell command. See ?cat for details.
3.5.4 Formatting Output
R provides a variety of ways to format data before you print it. Taking the time to format output carefully makes it easier to read and understand, as well as making your scripts seem more professional.
3.5.4.1 Escape Sequences
One way to format strings is by adding (or removing) escape sequences. An escape sequence is a sequence of characters that represents some other character, usually one that’s invisible (such as whitespace) or doesn’t appear on a standard keyboard.
In R, escape sequences always begin with a backslash. For example, \n is a
newline. The message and cat functions automatically convert escape
sequences to the characters they represent:
## Hello
## world!The print function doesn’t convert escape sequences:
## [1] "Hello\nworld!"Some escape sequences trigger special behavior in the console. For example,
ending a line with a carriage return \r makes the console print the next line
over the line. Try running this code in a console (it’s not possible to see the
result in a static book):
# Run this in an R console.
for (i in 1:10) {
message(i, "\r", appendLF = FALSE)
# Wait 0.5 seconds.
Sys.sleep(0.5)
}You can find a complete list of escape sequences in ?Quotes.
3.5.4.2 Formatting Functions
You can use the sprintf function to apply specific formatting to values and
substitute them into strings. The function uses a mini-language to describe the
formatting and substitutions. The sprintf function (or something like it) is
available in many programming languages, so being familiar with it will serve
you well on your programming journey.
The key idea is that substitutions are marked by a percent sign % and a
character. The character indicates the kind of data to be substituted: s for
strings, i for integers, f for floating point numbers, and so on.
The first argument to sprintf must be a string, and subsequent arguments are
values to substitute into the string (from left to right). For example:
## [1] "My age is 32, and my name is Nick"You can use the mini-language to do things like specify how many digits to
print after a decimal point. Format settings for a substituted value go between
the percent sign % and the character. For instance, here’s how to print pi
with 2 digits after the decimal:
## [1] "3.14"You can learn more by reading ?sprintf.
Much simpler are the paste and paste0 functions, which coerce their
arguments to strings and concatenate (or “paste together”) them. The paste
function inserts a space between each argument, while the paste0
function doesn’t:
## [1] "Hello world"## [1] "Helloworld"You can control the character inserted between arguments with the sep
parameter.
By setting an argument for the collapse parameter, you can also use the
paste and paste0 functions to concatenate the elements of a vector. The
argument to collapse is inserted between the elements. For example, suppose
you want to paste together elements of a vector inserting a comma and space
, in between:
## [1] "1, 2, 3"Members of the R community have developed many packages to make formatting strings easier:
3.5.5 Logging Output
Logging means saving the output from some code to a file as the code runs. The file where the output is saved is called a log file or log, but this name isn’t indicative of a specific format (unlike, say, a “CSV file”).
It’s a good idea to set up some kind of logging for any code that takes more than a few minutes to run, because then if something goes wrong you can inspect the log to diagnose the problem. Think of any output that’s not logged as ephemeral: it could disappear if someone reboots the computer, or there’s a power outage, or some other, unforeseen event.
R’s built-in tools for logging are rudimentary, but members of the community have developed a variety of packages for logging. Here are a few that are still actively maintained as of January 2023:
- logger – a relatively new package that aims to improve aspects of other logging packages that R users find confusing.
- futile.logger – a popular, mature logging package based on Apache’s Log4j utility and on R idioms.
- logging – a mature logging package based on Python’s
loggingmodule. - loggit – integrates with R’s conditions system and writes logs in JavaScript Object Notation (JSON) format so they are easy to inspect programmatically.
- log4r – another package based on Log4j with an object-oriented programming approach.
3.6 Reading Input
This section explains how to make R read inputs from the command line, configuration files, and interactive prompts. These kinds of inputs are often important for scripts, because they allow for flexibility in initial parameters and how run-time events are handled.
3.6.1 Command Line Arguments
When you run an R script from the command line, you can optionally pass additional arguments to the script by putting them after command. Command line arguments are a convenient and relatively safe way to set parameters in scripts. This is in contrast to editing scripts directly, which may be difficult (if the code is complex or disorganized) and may introduce bugs. That said, for scripts with lots of parameters, we recommend reading the parameters from a configuration file (see Section 3.6.2) instead or in addition to command line arguments.
You can capture command line arguments with R’s built-in the commandArgs
function or with functions from a variety of packages. By default, the
commandArgs function returns the entire command used to run R, including the
Rscript command and the name of the script. You can get just the arguments
after the name of the script by setting trailingOnly = TRUE. The function
assumes arguments are space-separated and returns a character vector with one
element for each argument. As an example, set up a script called
hello_name.R:
main = function(
args = commandArgs(trailingOnly = TRUE)
) {
if (length(args) < 1)
message("Usage: Rscript hello_name.R NAME")
message("Hello, ", args[[1]], "!")
}
main()Try running the script from the command line with this command:
Rscript hello_name.R Susan## Hello, Susan!Several packages on CRAN provide functions for more complex processing of command line arguments. As of 2024, many of these packages appear to be unmaintained or have undesirable dependencies (for example, the argparse package depends on Python). One of the more promising packages is optparse.
3.6.2 Configuration Files
When a script has lots of parameters, reading them from a separate configuration file is often a good idea. Configuration files are beneficial because they provide a record of parameter settings that can be saved separately from the code (with command line arguments, it’s easy to forget which settings you used in earlier runs).
There are many different formats available for configuration files. Two that are especially popular are:
Yet Another Markup Language (YAML) – YAML is a plain text format designed to be easy for people to read. It’s widely used in the R community. For instance, R Markdown notebooks typically include a YAML header for settings. The yaml package provides functions to read and write YAML files.
Tom’s Obvious Markup Language (TOML) – TOML is also a plain text format designed to be easy for people to read. Compared to YAML, TOML’s main advantage is that its syntax is simpler and thus easier to learn. TOML is widely used in other programming communities (especially Python and Rust), but is not yet widely used in the R community. The RcppTOML package provides functions to read TOML files.
3.6.3 Prompts
Sometimes it’s useful to prompt the user for input while code is running. For instance, a script that produces output can prompt the user about what to do if the output would overwrite an existing file. You can also use prompts to develop interactive programs, which may be more accessible to users unfamiliar with programming and command line interfaces.
One way to prompt the user for input is with R’s built-in readline function.
The function prompts the user for input and returns whatever they enter as a
string. Try out the readline function in an R prompt:
A more sophisticated way to prompt the user for input is with R’s built-in
scan function. The scan function attempts to read a specific type of data,
which you can customize. By default, the function prompts the user for numbers
and returns when they enter a blank line. The result is a vector with one
element for each number they entered. Try out the scan function in an R
prompt:
You can set the what parameter in scan to specify the type of data to read.
For instance, if you want to read strings (into a character vector), set what = character():
The scan function has many other parameters, which you can use to control
things like how many values the user can enter and which values should be
interpreted as the missing value NA.